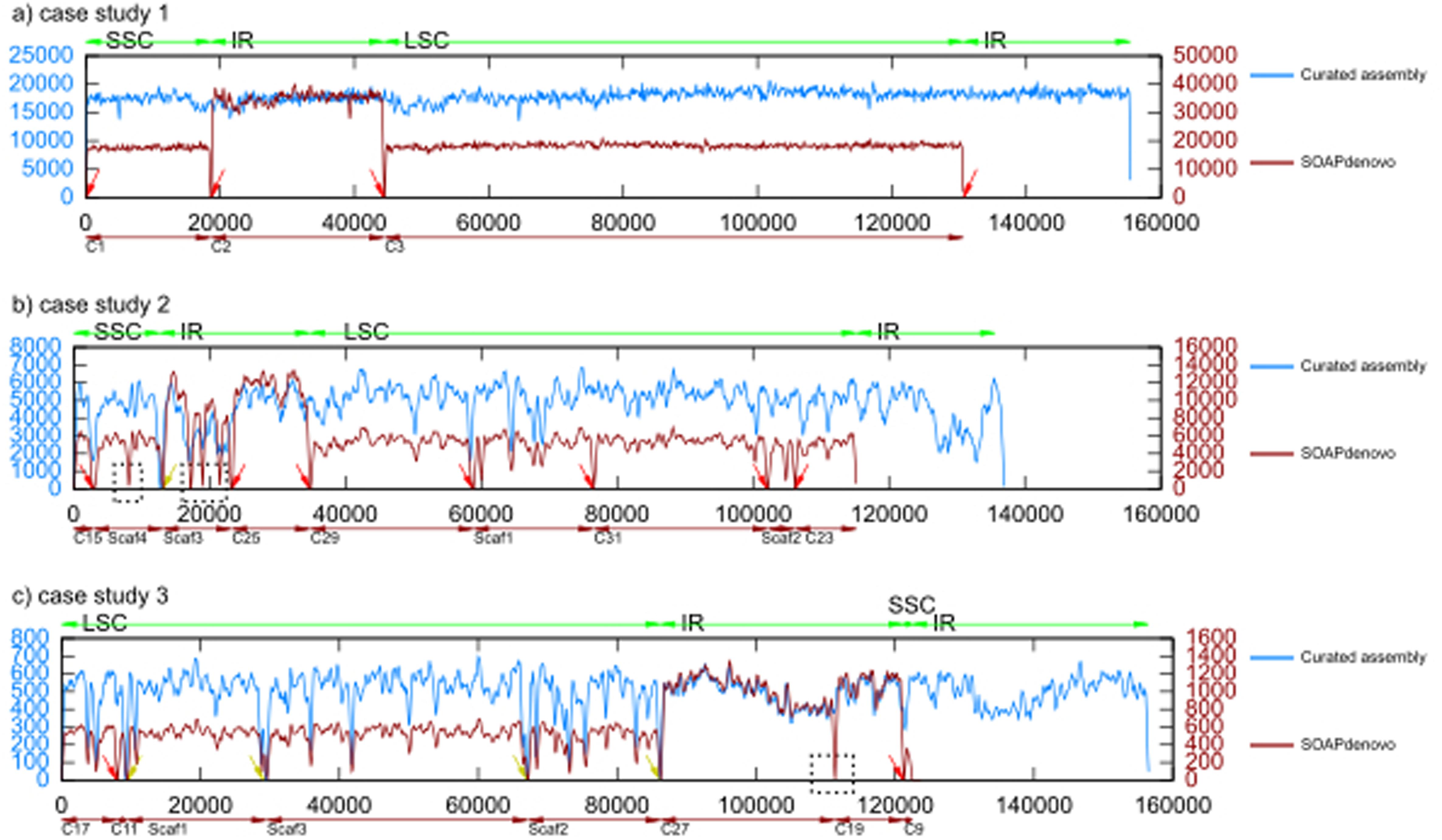
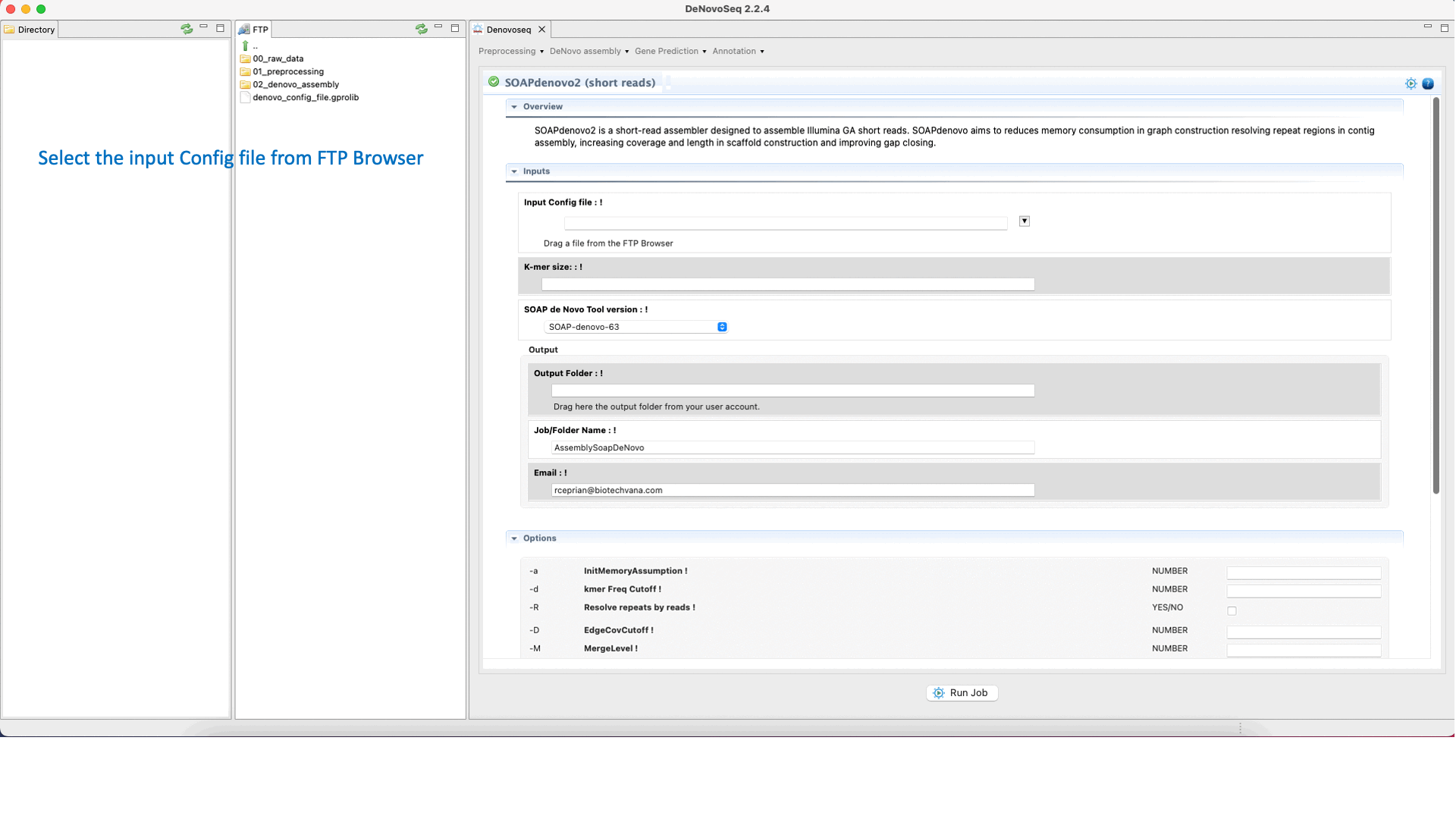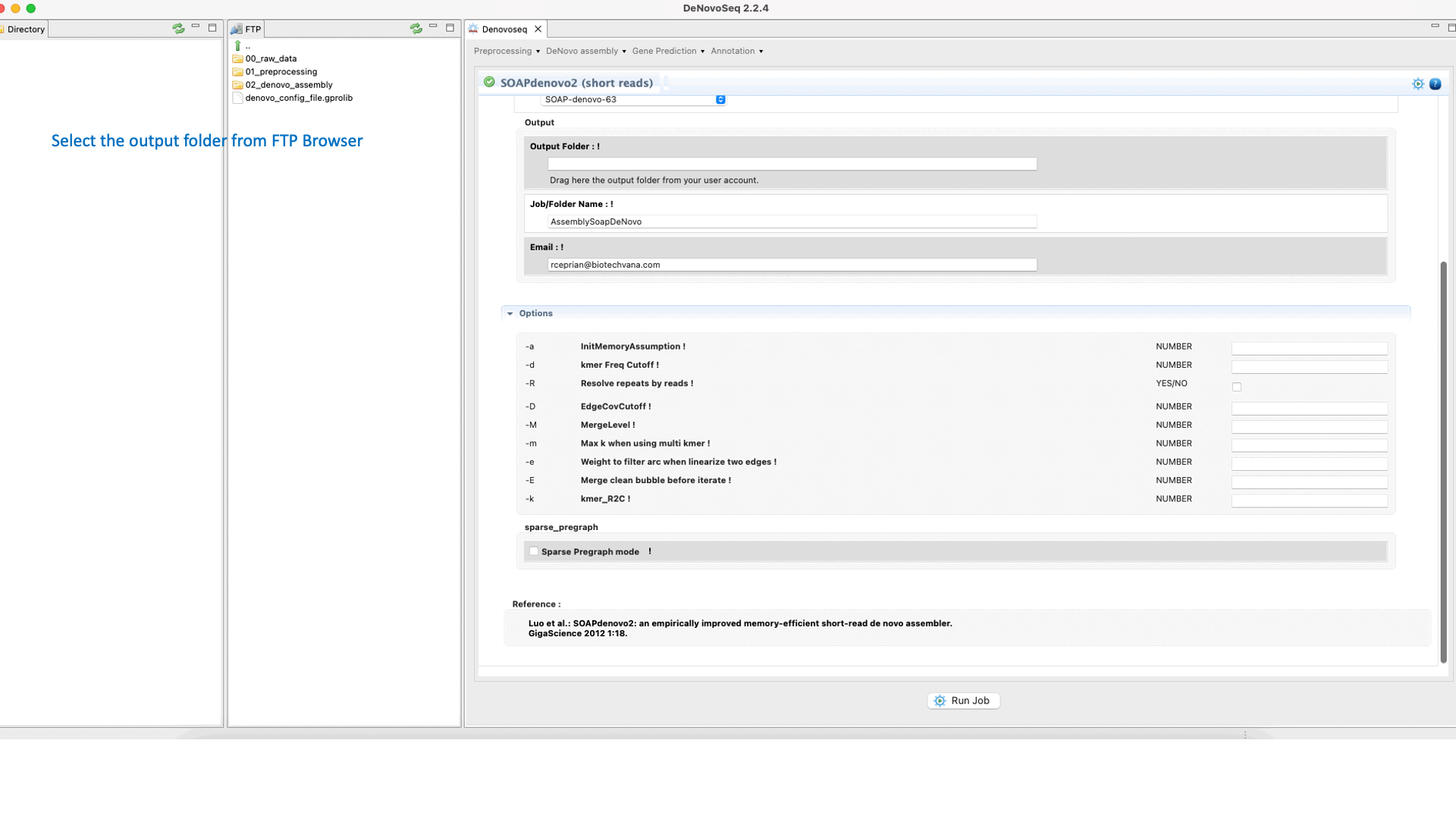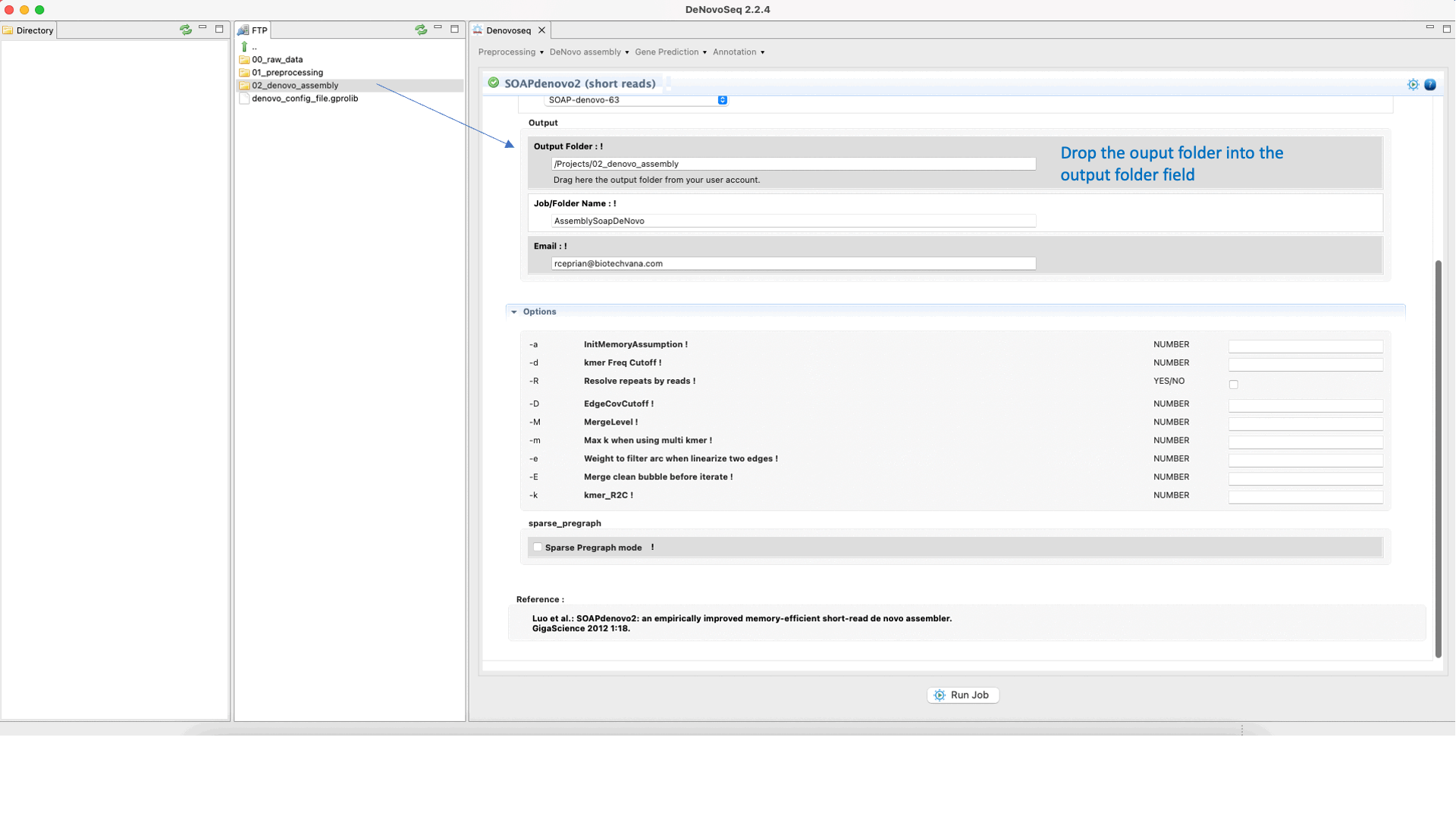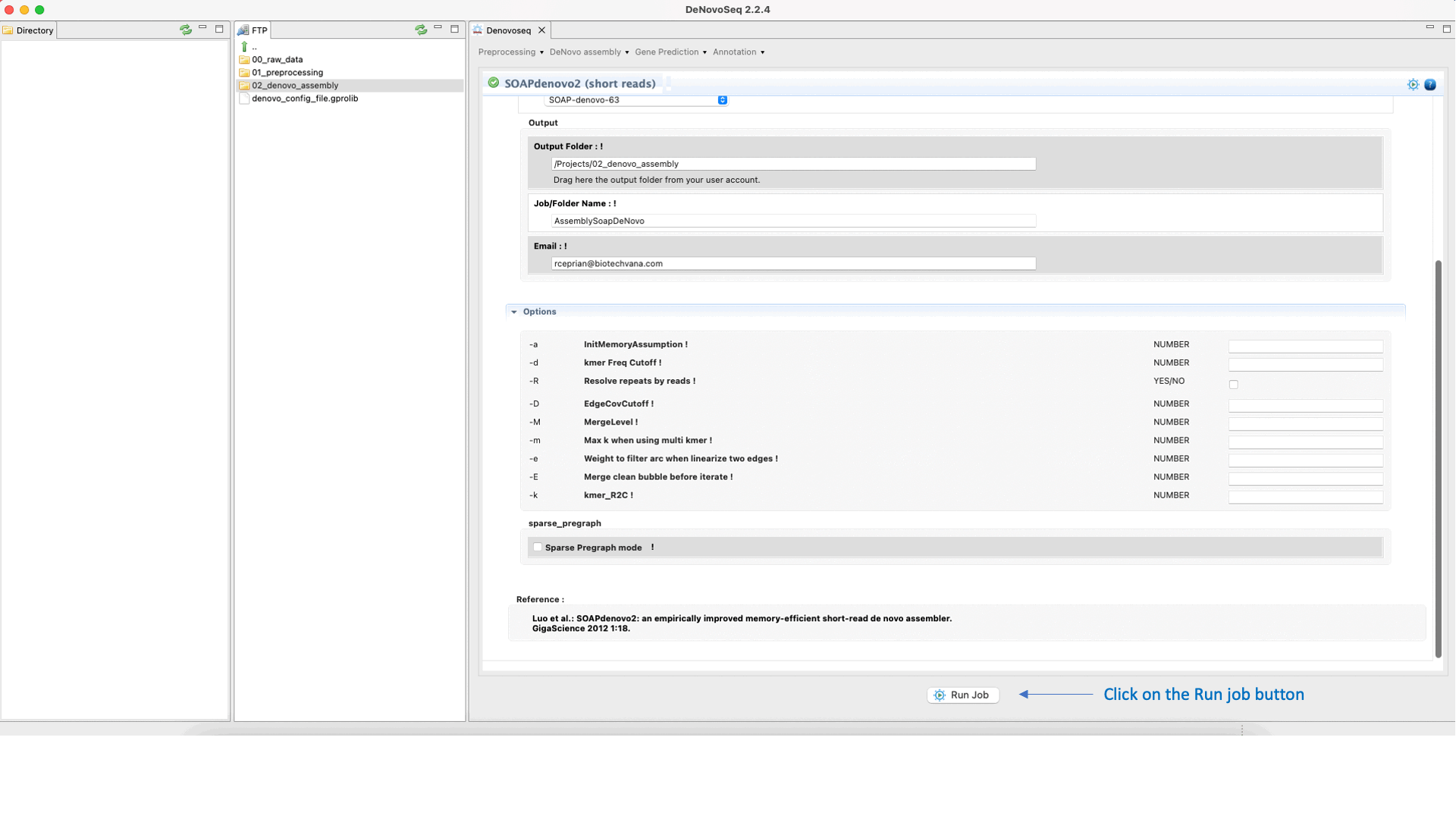
Contig continuity, genome and gene coverages for de novo assembly with... | Download Scientific Diagram

Overview of SOAPdenovo-Trans algorithm. (A1) Contig assembly. De Bruijn... | Download Scientific Diagram

Challenges and opportunities for strain verification by whole-genome sequencing | Scientific Reports

SCERNA Flowchart. SCERNA stands for Scaffolding and Error correction... | Download Scientific Diagram

Schematic workflow showing the de novo short read assembly approaches... | Download Scientific Diagram
Identification of Optimum Sequencing Depth Especially for De Novo Genome Assembly of Small Genomes Using Next Generation Sequencing Data | PLOS ONE

De novo transcriptome assembly using QIAGEN CLC Genomics Workbench - Bioinformatics Software | QIAGEN Digital Insights

The pipeline. Sixteen transcriptomes were assembled using both de novo... | Download Scientific Diagram

Reference-guided de novo assembly approach improves genome reconstruction for related species | BMC Bioinformatics | Full Text
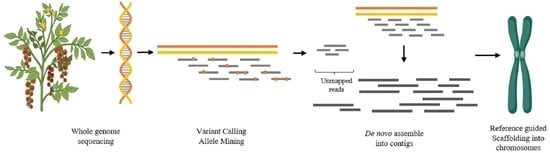
Genes | Free Full-Text | Reference Guided De Novo Genome Assembly of Transformation Pliable Solanum lycopersicum cv. Pusa Ruby

Figure S2. De novo assembly process using Velvet, SOAP de novo, and CLC... | Download Scientific Diagram
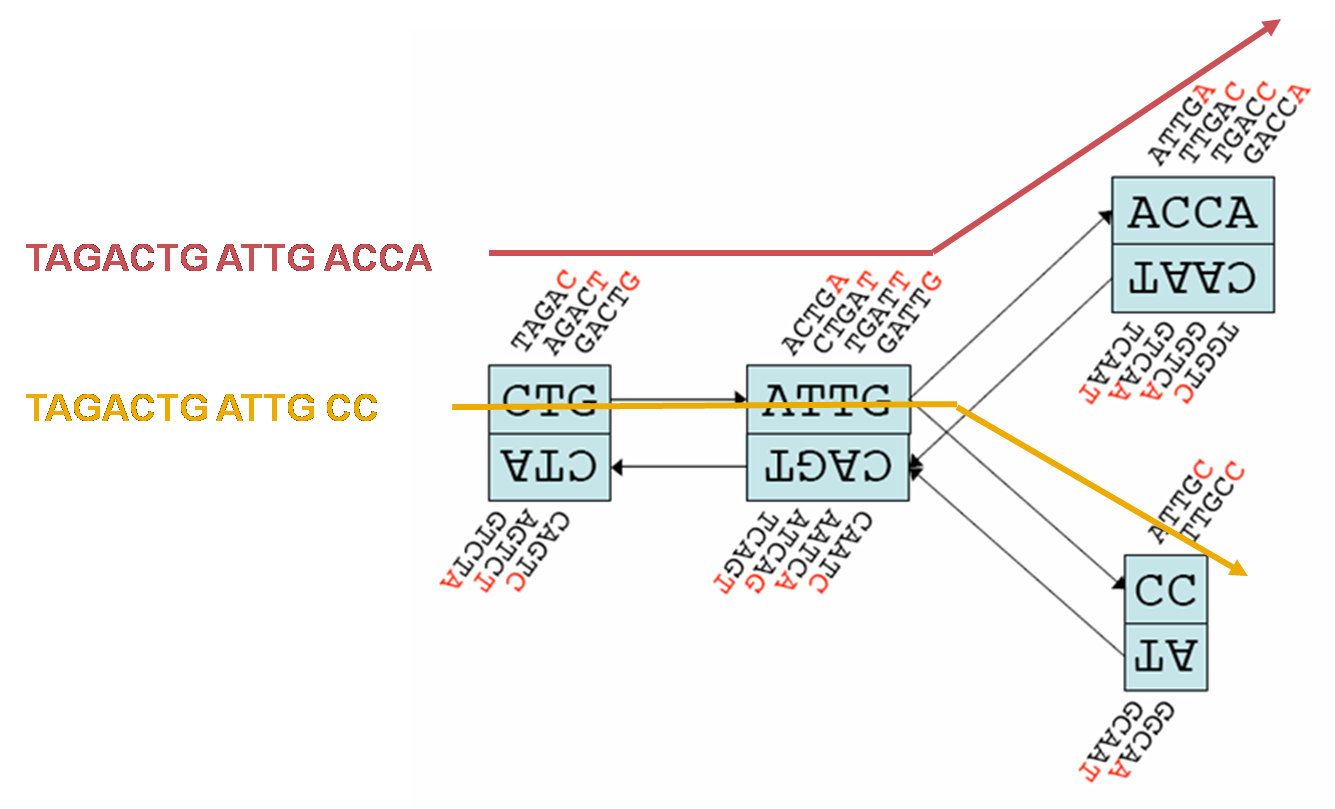
GitHub - ShujiaHuang/SOAPdenovo: SOAPdenovo is a novel short-read assembly method that can build a de novo draft assembly for the human-sized genomes.